Developers Guide#
Here we document internal components of Arbor. These pages can be useful if you’re interested in developing on Arbor itself.
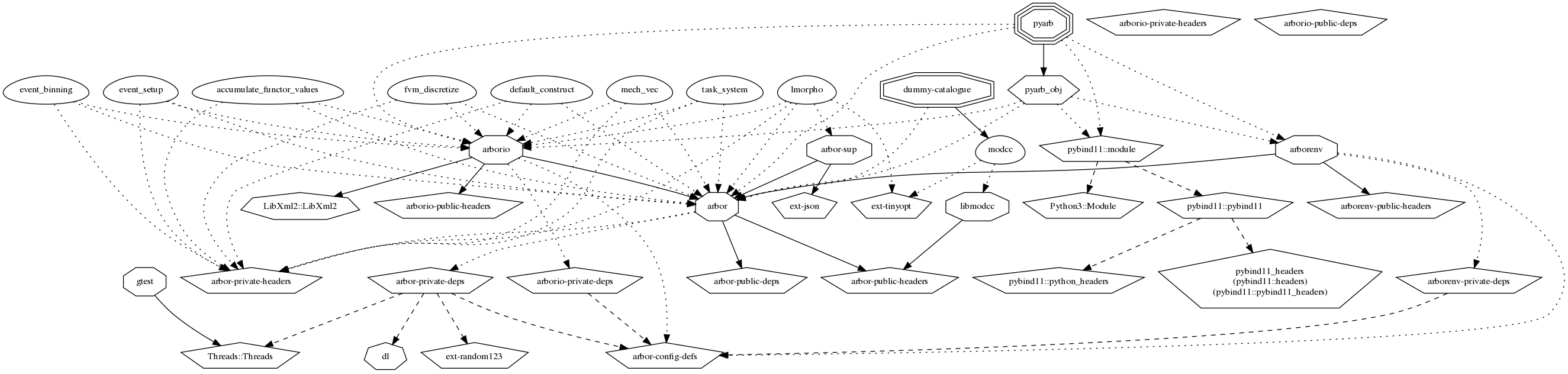
Arbor dependency graph. To see what dependencies Arbor has and how to generate this figure, see Dependency management.#
Arbor Developers Guide:
- How the Cable Cell is made
- Cell groups
- Communication in Arbor
- Exchange of Spikes
- Distribution of Events to Targets
- Building the Connection Table
- Debugging
- Matrix Solvers
- Stochastic Differential Equations
- SIMD Classes
- Shared State
- Exporting Symbols
- Adding Built-in Catalogues to Arbor
- Mechanism ABI
- Utility wrappers and containers
- Version and build information
- Numerical methods